December 4, 2019
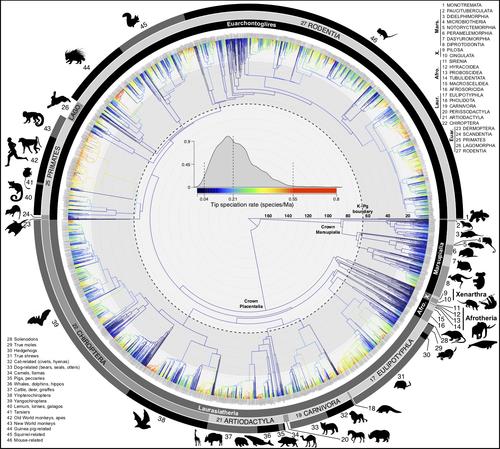
A new Jetz Lab paper, led by post-doc Nathan Upham, was published in PLOS Biology describing efforts to build an improved evolutionary tree of life for all ~6,000 species of living mammals. This paper was four-years in the making as part of the NSF VertLife Terrestrial grant, which has been focused on improving the species-level resolution of phylogenetic and trait data across the ~33,000 species of terrestrial vertebrates (tetrapods). Mammals were the last piece of that puzzle, with phylogenies of birds, squamates, and amphibians published in the last several years.
With the publication of the mammal trees comes some exciting advances in the ability of researchers to accurately investigate the ~180-million year history of crown mammals. Rather than relying on a single “best” consensus tree of life, levels of confidence in the evolutionary relationships, ages, and rates are now incorporated across 10,000 trees. Researchers can sample from this credible set of trees to better account for the inherent uncertainty in reconstructing deep-time historical events from DNA sequences and fossils. These data are often fragmentary and non-randomly sampled, so there is even greater need to be honest about reconstructions of past evolutionary dynamics.
Paper in PLoS Biology: https://doi.org/10.1371/journal.pbio.3000494
Explore the tree: http://vertlife.org/data/mammals/
Subset and download trees: http://vertlife.org/phylosubsets
Explanation on Twitter: https://twitter.com/n8_upham/status/1202334242392494080
